FAQ#
Answers to frequently asked questions about the cleanlab open-source package.
The code snippets in this FAQ come from a fully executable notebook you can run via Colab or locally by downloading it here.
What data can cleanlab detect issues in?#
Currently, cleanlab can be used to detect label issues in any classification dataset, including those involving: multiple annotators per example (multi-annotator), or multiple labels per example (multi-label). This includes data from any modality such as: image, text, tabular, audio, etc. For text data, cleanlab also supports NLP tasks like entity recognition in which each word is individually labeled (token classification). We’re working to add support for all other common supervised learning tasks. If you have a particular task in mind, let us know!
How do I format classification labels for cleanlab?#
With Datalab:
Datalab simplifies label management by accepting both string and integer labels directly. Internally, unique labels are sorted alphanumerically and mapped to integers, facilitating seamless integration with lower-level cleanlab methods. Below are the supported label formats:
List of strings or integers: Directly pass labels as a list of strings or integers without manual encoding.
Using
datasets.DatasetwithClassLabel: For advanced use cases, you can structure your dataset using HuggingFace’sdatasets.Datasetobject, specifying label columns asClassLabelfeature objects for formatting the labels. Refer to the datasets documentation for detailed guidance.
from cleanlab import Datalab
from datasets import Dataset, Features, Value, ClassLabel
# Example 1: Labels as a list of strings
labels_str = ['cat', 'dog', 'cat', 'dog']
datalab_str = Datalab(data={"text": ["a", "b", "c", "d"], "label": labels_str}, label_name="label")
print("String labels:", datalab_str.labels)
# Example 2: Labels as a list of integers
labels_int = [1, 2, 2, 1] # These will be remapped to [0, 1] internally
datalab_int = Datalab(data={"text": ["a", "b", "c", "d"], "label": labels_int}, label_name="label")
print("Integer labels:", datalab_int.labels)
# Example 3: Advanced - Dataset with ClassLabel feature
my_dict = {"pet_name": ["Spot", "Mittens", "Rover", "Rocky", "Pepper", "Socks"], "species": ["dog", "cat", "dog", "dog", "cat", "cat"]}
features = Features({"pet_name": Value("string"), "species": ClassLabel(names=["dog", "cat"])})
dataset = Dataset.from_dict(my_dict, features=features)
datalab_dataset = Datalab(data=dataset, label_name="species")
print("ClassLabel feature:", datalab_dataset.labels)
Using Datalab allows you to directly handle raw class name labels in your dataset while ensuring compatibility with label encoding requirements of lower-level cleanlab methods, which we’ll cover in the next section.
Without Datalab:
Outside of Datalab, cleanlab offers various lower-level methods to directly operate on labels and diagnose issues. For instance: get_label_quality_scores() and find_label_issues(). These lower-level methods only work with integer-encoded labels in the range {0,1, ... K-1} where K = number_of_classes. The labels array should only contain integer values in the range {0, K-1} and be of shape (N,) where N = total_number_of_data_points. Do not pass in labels where
some classes are entirely missing or are extremely rare, as cleanlab may not perform as expected. It is better to remove such classes entirely from the dataset first (also dropping the corresponding dimensions from pred_probs and then renormalizing it).
Text or string labels should to be mapped to integers for each possible value. For example if your original data labels look like this: ["dog", "dog", "cat", "mouse", "cat"], you should feed them to cleanlab like this: labels = [1,1,0,2,0] and keep track of which integer uniquely represents which class (classes were ordered alphabetically in this example).
One-hot encoded labels should be integer-encoded by finding the argmax along the one-hot encoded axis. An example of what this might look like is shown below.
[2]:
import numpy as np
# This example arr has 4 labels (one per data point) where
# each label can be one of 3 possible classes
arr = np.array([[0,1,0],[1,0,0],[0,0,1],[1,0,0]])
labels_proper_format = np.argmax(arr, axis=1) # How labels should be formatted when passed into the model
How do I infer the correct labels for examples cleanlab has flagged?#
If you have a classifier that is compatible with CleanLearning (i.e. follows the sklearn API), here’s an easy way to see predicted labels alongside the label issues:
[3]:
cl = cleanlab.classification.CleanLearning(your_classifier)
issues_dataframe = cl.find_label_issues(data, labels)
Alternatively if you have already computed out-of-sample predicted probabilities (pred_probs) from a classifier:
[4]:
cl = cleanlab.classification.CleanLearning()
issues_dataframe = cl.find_label_issues(X=None, labels=labels, pred_probs=pred_probs)
Otherwise if you have already found issues via:
[5]:
issues = cleanlab.filter.find_label_issues(labels, pred_probs)
then you can see your trained classifier’s class prediction for each flagged example like this:
[6]:
class_predicted_for_flagged_examples = pred_probs[issues].argmax(axis=1)
Here you can see the classifier’s class prediction for every example via:
[7]:
class_predicted_for_all_examples = pred_probs.argmax(axis=1)
We caution against just blindly taking the predicted label for granted, many of these suggestions may be wrong! You will be able to produce a much better version of your dataset interactively using Cleanlab Studio, which helps you efficiently fix issues like this in large datasets.
How should I handle label errors in train vs. test data?#
If you do not address label errors in your test data, you may not even know when you have produced a better ML model because the evaluation is too noisy. For the best-trained models and most reliable evaluation of them, you should fix label errors in both training and testing data.
To do this efficiently, first use cleanlab to automatically find label issues in both sets. You can simply merge these two sets into one larger dataset and run cross-validation training. On the merged dataset, you can do either of the following to detect label issues:
With Datalab: Run Datalab.find_issues() on the merged dataset, then call Datalab.report() to see the label issues (and other types of data issues).
from cleanlab import Datalab
lab = Datalab(data = merged_dataset, label_name = "label_column_name")
# Run proper cross-validation when computing predicted probabilities
lab.find_issues(pred_probs = pred_probs, issue_types = {"label": {}})
lab.report()
You can fetch the label issues DataFrame from the Datalab object by calling:
label_issues = lab.get_issues("label")
Without Datalab: Run cleanlab’s lower-level find_label_issues() method on the merged datataset. Calling the CleanLearning.find_label_issues() method on your merged dataset both runs cross-validation training and finds label issues for you with any scikit-learn compatible classifier you choose.
After finding label issues, be wary about auto-correcting the labels for test examples. Instead manually fix the labels for your test data via careful review of the flagged issues. You can use Cleanlab Studio to fix labels efficiently.
Auto-correcting labels for your training data is fair game, which should improve ML performance (if properly evaluated with clean test labels). You can boost ML performance further by manually fixing the training examples flagged with label issues, as demonstrated in this article:
Handling Mislabeled Tabular Data to Improve Your XGBoost Model
How can I find label issues in big datasets with limited memory?#
For a dataset with many rows and/or classes, there are more efficient methods in the label_issues_batched module. These methods read data in mini-batches and you can reduce the batch_size to control how much memory they require. Below is an example of how to use the find_label_issues_batched() method from this module, which can load mini-batches of data from labels, pred_probs saved as .npy files on disk. You can also run this method on Zarr arrays loaded from .zarr files.
Try playing with the n_jobs argument for further multiprocessing speedups. If you need greater flexibility, check out the LabelInspector class from this module.
[8]:
# We'll assume your big arrays of labels, pred_probs have been saved to file like this:
from tempfile import mkdtemp
import os.path as path
labels_file = path.join(mkdtemp(), "labels.npy")
pred_probs_file = path.join(mkdtemp(), "pred_probs.npy")
np.save(labels_file, labels)
np.save(pred_probs_file, pred_probs)
# Code to find label issues by loading data from file in batches:
from cleanlab.experimental.label_issues_batched import find_label_issues_batched
batch_size = 10000 # for efficiency, set this to as large of a value as your memory can handle
# Indices of examples with label issues, sorted by label quality score (most severe to least severe):
indices_of_examples_with_issues = find_label_issues_batched(
labels_file=labels_file, pred_probs_file=pred_probs_file, batch_size=batch_size
)
mmap-loaded numpy arrays have: 50 examples, 3 classes
Total number of examples whose labels have been evaluated: 50
Methods that internally call filter.find_label_issues() can be sped up by specifying the argument low_memory=True, which will instead use find_label_issues_batched() internally. The following methods provide this option:
To use less memory and get results faster if your dataset has many classes: Try merging the rare classes into a single “Other” class before you find label issues. The resulting issues won’t be affected much since cleanlab anyway does not have enough data to accurately diagnose label errors in classes that are rarely seen. To do this, you should aggregate all the probability assigned to the rare classes in pred_probs into a single new dimension of pred_probs_merged (where this new
array no longer has columns for the rare classes). Here is a function that does this for you, which you can also modify as needed:
[11]:
from cleanlab.internal.util import value_counts # use this to count how often each class occurs in labels
def merge_rare_classes(labels, pred_probs, count_threshold = 10):
"""
Returns: labels, pred_probs after we merge all rare classes into a single 'Other' class.
Merged pred_probs has less columns. Rare classes are any occuring less than `count_threshold` times.
Also returns: `class_mapping_orig2new`, a dict to map new classes in merged labels back to classes
in original labels, useful for interpreting outputs from `dataset.heath_summary()` or `count.confident_joint()`.
"""
num_classes = pred_probs.shape[1]
num_examples_per_class = value_counts(labels, num_classes=num_classes)
rare_classes = [c for c in range(num_classes) if num_examples_per_class[c] < count_threshold]
if len(rare_classes) < 1:
raise ValueError("No rare classes found at the given `count_threshold`, merging is unnecessary unless you increase it.")
num_classes_merged = num_classes - len(rare_classes) + 1 # one extra class for all the merged ones
other_class = num_classes_merged - 1
labels_merged = labels.copy()
class_mapping_orig2new = {} # key = original class in `labels`, value = new class in `labels_merged`
new_c = 0
for c in range(num_classes):
if c in rare_classes:
class_mapping_orig2new[c] = other_class
else:
class_mapping_orig2new[c] = new_c
new_c += 1
labels_merged[labels == c] = class_mapping_orig2new[c]
merged_prob = np.sum(pred_probs[:, rare_classes], axis=1, keepdims=True) # total probability over all merged classes for each example
pred_probs_merged = np.hstack((np.delete(pred_probs, rare_classes, axis=1), merged_prob)) # assumes new_class is as close to original_class in sorted order as is possible after removing the merged original classes
# check a few rows of probabilities after merging to verify they still sum to 1:
num_check = 1000 # only check a few rows for efficiency
ones_array_ref = np.ones(min(num_check,len(pred_probs)))
if np.isclose(np.sum(pred_probs[:num_check], axis=1), ones_array_ref).all() and (not np.isclose(np.sum(pred_probs_merged[:num_check], axis=1), ones_array_ref).all()):
raise ValueError("merged pred_probs do not sum to 1 in each row, check that merging was correctly done.")
return (labels_merged, pred_probs_merged, class_mapping_orig2new)
[12]:
from cleanlab.filter import find_label_issues # can alternatively use find_label_issues_batched() shown above
labels_merged, pred_probs_merged, class_mapping_orig2new = merge_rare_classes(labels, pred_probs, count_threshold=5)
examples_w_issues = find_label_issues(labels_merged, pred_probs_merged, return_indices_ranked_by="self_confidence")
Why isn’t CleanLearning working for me?#
At this time, CleanLearning only works with data formatted as numpy matrices or pd.DataFrames, and with models that are compatible with the sklearn API (check out skorch for Pytorch compatibility and scikeras for Tensorflow/Keras compatibility). You can still use cleanlab with other data formats though! Just separately obtain predicted probabilities (pred_probs) from your model via cross-validation and
pass them as inputs.
If CleanLearning is running successfully but not improving predictive accuracy of your model, here are some tips:
Use cleanlab to find label issues in your test data as well (we recommend pooling
labelsacross both training and test data into one input forfind_label_issues()). Then manually review and fix label issues identified in the test data to verify accuracy measurements are actually meaningful.Try different values for
filter_by,frac_noise, andmin_examples_per_classwhich can be set via thefind_label_issues_kwargsargument in the initialization ofCleanLearning().Try to find a better model (eg. via hyperparameter tuning or changing to another classifier).
CleanLearningcan find better label issues by leveraging a better model, which allows it to produce better quality training data. This can form a virtuous cycle in which better models -> better issue detection -> better data -> even better models!Try jointly tuning both model hyperparameters and
find_label_issues_kwargsvalues.Does your dataset have a junk (or clutter, unknown, other) class? If you have bad data, consider creating one (c.f. Caltech-256).
Consider merging similar/overlapping classes found via
cleanlab.dataset.find_overlapping_classes.
Other general tips to improve label error detection performance:
Try creating more restrictive new filters by combining their intersections (e.g.
combined_boolean_mask = mask1 & mask2wheremask1andmask2are the boolean masks created by runningfind_label_issueswith different values of thefilter_byargument).If your
pred_probsare obtained via a neural network, try averaging thepred_probsover the last K epochs of training instead of just using the finalpred_probs. Similarly, you can try averagingpred_probsfrom several models (remember to re-normalize) or usingcleanlab.rank.get_label_quality_ensemble_scores.
How can I use different models for data cleaning vs. final training in CleanLearning?#
The code below demonstrates CleanLearning with 2 different classifiers: LogisticRegression() and GradientBoostingClassifier(). A LogisticRegression model is used to detect label issues (via cross-validation run inside CleanLearning) and a GradientBoostingClassifier model is finally trained on a clean subset of the data with issues removed. This can be done with any two classifiers.
[14]:
from cleanlab.classification import CleanLearning
import numpy as np
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import GradientBoostingClassifier
# Make example data
data = np.vstack([np.random.random((100, 2)), np.random.random((100, 2)) + 10])
labels = np.array([0] * 100 + [1] * 100)
# Introduce label errors
true_errors = [97, 98, 100, 101, 102, 104]
for idx in true_errors:
labels[idx] = 1 - labels[idx]
# CleanLearning with 2 different classifiers: one classifier is used to detect label issues
# and a different classifier is subsequently trained on the clean subset of the data.
model_to_find_errors = LogisticRegression() # this model will be trained many times via cross-validation
model_to_return = GradientBoostingClassifier() # this model will be trained once on clean subset of data
cl0 = CleanLearning(model_to_find_errors)
issues = cl0.find_label_issues(data, labels)
cl = CleanLearning(model_to_return).fit(data, labels, label_issues=issues)
pred_probs = cl.predict_proba(data) # predictions from GradientBoostingClassifier
print(cl0.clf) # will be LogisticRegression()
print(cl.clf) # will be GradientBoostingClassifier()
LogisticRegression()
GradientBoostingClassifier()
How do I hyperparameter tune only the final model trained (and not the one finding label issues) in CleanLearning?#
The code below demonstrates CleanLearning using a GradientBoostingClassifier() with no hyperparameter-tuning to find label issues but with hyperparameter-tuning via RandomizedSearchCV(...) for the final training of this model on the clean subset of the data. This is a useful trick to avoid expensive hyperparameter-tuning for every fold of cross-validation (which is needed to find label issues).
[15]:
import numpy as np
from cleanlab.classification import CleanLearning
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import RandomizedSearchCV
# Make example data
data = np.vstack([np.random.random((100, 2)), np.random.random((100, 2)) + 10])
labels = np.array([0] * 100 + [1] * 100)
# Introduce label errors
true_errors = [97, 98, 100, 101, 102, 104]
for idx in true_errors:
labels[idx] = 1 - labels[idx]
# CleanLearning with no hyperparameter-tuning during expensive cross-validation to find label issues
# but hyperparameter-tuning for the final training of model on clean subset of the data:
model_to_find_errors = GradientBoostingClassifier() # this model will be trained many times via cross-validation
model_to_return = RandomizedSearchCV(GradientBoostingClassifier(),
param_distributions = {
"learning_rate": [0.001, 0.05, 0.1, 0.2, 0.5],
"max_depth": [3, 5, 10],
}
) # this model will be trained once on clean subset of data
cl0 = CleanLearning(model_to_find_errors)
issues = cl0.find_label_issues(data, labels)
cl = CleanLearning(model_to_return).fit(data, labels, label_issues=issues) # CleanLearning for hyperparameter final training
pred_probs = cl.predict_proba(data) # predictions from hyperparameter-tuned GradientBoostingClassifier
print(cl0.clf) # will be GradientBoostingClassifier()
print(cl.clf) # will be RandomizedSearchCV(estimator=GradientBoostingClassifier(),...)
GradientBoostingClassifier()
RandomizedSearchCV(estimator=GradientBoostingClassifier(),
param_distributions={'learning_rate': [0.001, 0.05, 0.1, 0.2,
0.5],
'max_depth': [3, 5, 10]})
Why does regression.learn.CleanLearning take so long?#
To effectively identify errors in a regression dataset, the methods in regression.learn.CleanLearning estimate each datapoint’s aleatoric uncertainty (by fitting a second copy of the regression model to predict the residuals’ magnitudes), as well as its epistemic uncertainty (by fitting multiple copies of the regression model with bootstrap resampling). These uncertainty estimates help provide a robust quality score that accounts for the model’s imperfect predictions.
These uncertainty estimates help produce better results but require longer runtimes. Here are a few options to speed up the runtime of these methods:
Reduce the number of bootstrap resampling rounds by decreasing the
n_bootargument (default value is 5, set it to 0 to skip the epistemic uncertainty estimation entirely).Set
include_aleatoric_uncertainty=Falseto skip the aleatoric uncertainty estimation.Include less elements in the
coarse_search_rangeargument of regression.learn.CleanLearning.find_label_issues. This is overall set of values initially considered for estimating the fraction of data that have label issues.Reduce the
fine_search_sizeargument of regression.learn.CleanLearning.find_label_issues. A higher number represents a more thorough search to precisely estimate the fraction of data that have label issues.
Below is sample code on how to pass in these arguments.
[16]:
from cleanlab.regression.learn import CleanLearning
X = np.random.random(size=(30, 3))
coefficients = np.random.uniform(-1, 1, size=3)
y = np.dot(X, coefficients) + np.random.normal(scale=0.2, size=30)
# passing optinal arguments to reduce runtime
cl = CleanLearning(n_boot=1, include_aleatoric_uncertainty=False)
cl.find_label_issues(X, y, coarse_search_range=[0.05, 0.1], fine_search_size=2)
# you can also pass coarse_search_range and fine_search_size as kwargs to CleanLearning.fit
cl.fit(X, y, find_label_issues_kwargs={"coarse_search_range": [0.05, 0.1], "fine_search_size": 2})
[16]:
CleanLearning(include_aleatoric_uncertainty=False, model=LinearRegression(),
n_boot=1)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
CleanLearning(include_aleatoric_uncertainty=False, model=LinearRegression(),
n_boot=1)LinearRegression()
LinearRegression()
With Datalab:
Datalab runs CleanLearning under the hood when looking for label issues in regression datasets. Here’s how you can achieve the same behavior as calling CleanLearning.find_label_issues() in the code above using Datalab:
[17]:
from cleanlab import Datalab
lab = Datalab(data = {"X": X, "y": y}, label_name = "y", task="regression")
issue_types = {
"label": {
"clean_learning_kwargs": {"n_boot": 1, "include_aleatoric_uncertainty": False},
"coarse_search_range": [0.05, 0.1],
"fine_search_size": 2,
},
}
lab.find_issues(features=X, issue_types = issue_types)
Finding label issues ...
Audit complete. 3 issues found in the dataset.
How do I specify pre-computed data slices/clusters when detecting the Underperforming Group Issue?#
When detecting underperforming groups in a dataset, Datalab provides the option for passing pre-computed cluster IDs to find_issues. These cluster IDs can be obtained by grouping the features using any clustering algorithm of your choice (E.g. K-Means, DBSCAN, HDBSCAN etc). By default, Datalab will detect the underperforming group using the DBSCAN clustering algorithm.
Below is sample code on how to generate cluster IDs and pass them to find_issues:
[18]:
import numpy as np
from cleanlab import Datalab
from sklearn.cluster import KMeans
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_predict
# Make example data
features = np.vstack([np.random.random((100, 2)), np.random.random((100, 2)) + 10])
labels = np.array([0] * 100 + [1] * 100)
# Train classifier and generate out-of-sample probabilities
model = LogisticRegression()
pred_probs = cross_val_predict(model, features, labels, method="predict_proba")
# Group features into 8 clusters
clusterer = KMeans(n_init='auto', n_clusters=5)
cluster_ids = clusterer.fit_predict(features)
# Find underperforming group
lab = Datalab(data={"features": features, "y": labels}, label_name="y")
issue_types = {"underperforming_group": {"cluster_ids": cluster_ids}}
lab.find_issues(features=features, pred_probs=pred_probs, issue_types=issue_types)
Finding underperforming_group issues ...
Audit complete. 0 issues found in the dataset.
For a tabular dataset, you can alternatively use a categorical column’s values as cluster IDs:
[19]:
import pandas as pd
# Make tabular dataset with 1 continuous column and 1 categorical column
continuous_column = np.concatenate([np.random.random(100), np.random.random(100) + 10])
categorical_column = np.concatenate([np.random.randint(0, 2, 100), np.random.randint(1, 3, 100)])
labels = np.array([0] * 100 + [1] * 100)
data_df = pd.DataFrame({"Feature_A": continuous_column, "Feature_B": categorical_column, "labels": labels})
# Train classifier and generate out-of-sample probabilities
model = LogisticRegression()
features = data_df[["Feature_A", "Feature_B"]].to_numpy()
pred_probs = cross_val_predict(model, features, labels, method="predict_proba")
# Find underperforming group
lab = Datalab(data=data_df, label_name="labels")
issue_types = {"underperforming_group": {"cluster_ids": data_df["Feature_B"].values}}
lab.find_issues(features=features, pred_probs=pred_probs, issue_types=issue_types)
Finding underperforming_group issues ...
Audit complete. 0 issues found in the dataset.
How to handle near-duplicate data identified by cleanlab?#
cleanlab may identify near-duplicate examples in your dataset, these are examples that are very similar to each other and can potentially cause issues in model training and analytics. When near-duplicates are present, models may unexpectedly emphasize these examples, especially if they were accidentally duplicated. In such cases, it is crucial to remove the (near) duplicate copies from your dataset to ensure accurate and reliable results. A common strategy is to remove all but one of the
duplicates from your dataset. Here’s how you can achieve this with results from cleanlab’s Datalab class:
[20]:
from typing import Callable
import pandas as pd
def merge_duplicate_sets(df, merge_key: str):
"""Generate group keys for each row, then merge intersecting sets.
:param df: DataFrame with columns 'is_near_duplicate_issue' and 'near_duplicate_sets'
:param merge_key: Name of the column to store the merged sets
"""
df[merge_key] = df.apply(construct_group_key, axis=1)
merged_sets = consolidate_sets(df[merge_key].tolist())
df[merge_key] = df[merge_key].map(
lambda x: next(s for s in merged_sets if x.issubset(s))
)
return df
def construct_group_key(row):
"""Convert near_duplicate_sets into a frozenset and include the row's own index."""
return frozenset(row['near_duplicate_sets']).union({row.name})
def consolidate_sets(sets_list):
"""Merge sets if they intersect."""
# Convert the input list of frozensets to a list of mutable sets
sets_list = [set(item) for item in sets_list]
# A flag to keep track of whether any sets were merged in the current iteration
merged = True
# Continue the merging process as long as we have merged some sets in the previous iteration
while merged:
merged = False
new_sets = []
# Iterate through each set in our list
for current_set in sets_list:
# Skip empty sets
if not current_set:
continue
# Find all sets that have an intersection with the current set
intersecting_sets = [s for s in sets_list if s & current_set]
# If more than one set intersects, set the merged flag to True
if len(intersecting_sets) > 1:
merged = True
# Merge all intersecting sets into one set
merged_set = set().union(*intersecting_sets)
new_sets.append(merged_set)
# Empty the sets we've merged to prevent them from being processed again
for s in intersecting_sets:
sets_list[sets_list.index(s)] = set()
# Replace the original sets list with the new list of merged sets
sets_list = new_sets
# Convert the merged sets back to frozensets for the output
return [frozenset(item) for item in sets_list]
def lowest_score_strategy(sub_df):
"""Keep the row with the lowest near_duplicate_score."""
return sub_df['near_duplicate_score'].idxmin()
def filter_near_duplicates(data: pd.DataFrame, strategy_fn: Callable = lowest_score_strategy, **strategy_kwargs):
"""
Given a dataframe with columns 'is_near_duplicate_issue' and 'near_duplicate_sets',
return a series of boolean values where True indicates the rows to be removed.
The strategy_fn determines which rows to keep within each near_duplicate_set.
:param data: DataFrame with is_near_duplicate_issue and near_duplicate_sets columns
:param strategy_fn: Function to determine which rows to keep within each near_duplicate_set
:return: Series of boolean values where True indicates rows to be removed.
"""
# Filter out rows where 'is_near_duplicate_issue' is True to get potential duplicates
duplicate_rows = data.query("is_near_duplicate_issue").copy()
# Generate group keys for each row and merge intersecting sets
group_key = "sets"
duplicate_rows = merge_duplicate_sets(duplicate_rows, merge_key=group_key)
# Use the strategy function to determine the indices of the rows to keep for each group
to_keep_indices = duplicate_rows.groupby(group_key).apply(strategy_fn, **strategy_kwargs).explode().values
# Produce a boolean series indicating which rows should be removed
to_remove = ~data.index.isin(to_keep_indices)
return to_remove
The functions above collect sets of near-duplicate examples. Within each collection, a single example is chosen to be kept in the dataset. The rest of the examples in the collection are removed. Examples that are not near-duplicates of any other examples are kept in the dataset as well.
The choice of which example to keep in each set of near-duplicate examples can be made in a variety of ways. Here, the example with the lowest near-duplicate score is chosen. You can use any strategy that best suits your application by defining the strategy as a function and passing it as the strategy_fn argument to filter_near_duplicates(). Below is an example of how this is applied to a dataset.
[21]:
from cleanlab import Datalab
import numpy as np
# Assume you have a dataset with a set of 3 near-duplicate examples
features = np.random.random(size=(15, 3))
for neighbor in range(1, 3):
# Make examples 0, 1, and 2 near-duplicates of each other
features[neighbor] = features[0] + np.random.normal(scale=0.001, size=3)
# Identify near-duplicate examples with Datalab
your_dataset = {
"features": features,
}
lab = Datalab(data=your_dataset)
lab.find_issues(features = features, issue_types={"near_duplicate": {}})
# Pick out ids of near-duplicate examples to remove
near_duplicate_issues = (
lab.get_issues("near_duplicate")
.query("is_near_duplicate_issue")
.sort_values("near_duplicate_score")
)
ids_to_remove_series = filter_near_duplicates(near_duplicate_issues)
Finding near_duplicate issues ...
Audit complete. 3 issues found in the dataset.
/tmp/ipykernel_5754/1995098996.py:88: DeprecationWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
to_keep_indices = duplicate_rows.groupby(group_key).apply(strategy_fn, **strategy_kwargs).explode().values
[22]:
print("Near-duplicate examples to keep:", np.where(~ids_to_remove_series)[0].tolist())
print("Near-duplicate examples to remove:", np.where(ids_to_remove_series)[0].tolist())
Near-duplicate examples to keep: [0]
Near-duplicate examples to remove: [1, 2]
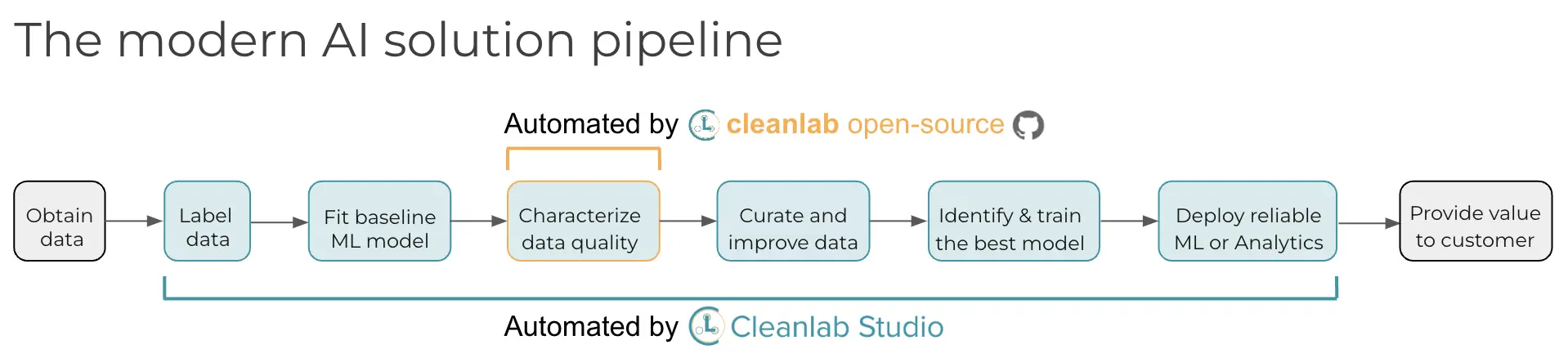
What ML models should I run cleanlab with? How do I fix the issues cleanlab has identified?#
These questions are automatically handled for you in Cleanlab Studio – our platform for no-code data improvement. While this open-source library finds data issues, an interface is needed to efficiently fix these issues in your dataset. Cleanlab Studio is a no-code platform to find and fix problems in real-world ML datasets. Cleanlab Studio automatically runs the data quality algorithms from this library on top of AutoML models fit to your data, and presents detected issues in a smart data editing interface. Think of it like a data cleaning assistant that helps you quickly improve the quality of your data (via AI/automation + streamlined UX). Try it for free!
What license is cleanlab open-sourced under?#
What does this mean? If you’re working at a company, you can use this open-source library to clean up your internal datasets. You can also use this open-source library to clean up a dataset used to train a model that is deployed in a commercial product. For non-commercial purposes, feel free to release altered versions of the source code as long as you include the same license.
Please email team@cleanlab.ai to discuss licensing needs if you would like to offer a commercial product that utilizes any cleanlab source code.
Can’t find an answer to your question?#
If your question is not addressed in these tutorials, please refer to the: Cleanlab Github issues, Cleanlab Code Examples or our Slack Community.
If your question is not addressed anywhere, please open a new Github issue. Our developers may also provide personalized assistance in our Slack Community.
Professional support and services are also available from our ML experts, learn more by emailing: team@cleanlab.ai